1. Introduction: The Project and Its Objectives
Overall Picture of the Project
Synthetic biology is a rapidly evolving field, and within it, iGEM serves as a gathering place for new ideas and research. Given its mission to solve various societal issues using synthetic biology, participation from individuals specializing in diverse fields is desirable. However, participating in this competition naturally requires extensive specialized knowledge in life sciences. In particular, the process of selecting Biobricks, the “components” of genes, can be very complex for beginners. To assist with this selection process, Dry-lab has developed a chatbot called “iGEM Copilot.”
Current Issues
The above represents one of the parts distributed this year. Additionally, parts with about this much information make up over 70% of the total. To be honest, the current iGEM registry often feels difficult to navigate for beginners. Specialized terminology is frequently used, and connections between these terms are lacking, making it challenging to follow the flow of information. This situation can potentially cause confusion and uncertainty during the information-seeking process. The purpose of iGEM Copilot is to solve these issues and make access to information easier for users.
What iGEM Copilot Aims to Achieve
The main objective of iGEM Copilot is to make the selection of Biobricks simpler and more efficient. This system is targeted especially at individuals who are participating in the complex field of iGEM for the first time. We have employed natural language processing technology to allow users to ask questions in the everyday language they are accustomed to using. This enables people who are not familiar with specialized terminology to easily find out what they want to do and what they need.
2. System Details
Natural Language-Based Component Recommendations
In response to specific questions like “Is there a way to store data using bacterial DNA?”, iGEM Copilot suggests the most suitable Biobricks. This feature is made possible by advanced machine learning algorithms. These algorithms analyze the user’s questions in natural language and quickly identify the most appropriate Biobricks by cross-referencing with the extensive iGEM registry.
Interactive User Interface
If users have any uncertainties or additional questions about the answers, they can ask on the spot and get immediate resolutions. This eliminates the need for users to search through multiple specialized databases or websites, saving them time and effort.
3. Technical Implementation and Evaluation
Comparison between Fine-tuning and Embedding
Two main methods for customizing language models are Fine-tuning and Embedding. This project adopted Embedding. The reason for this choice is that Embedding can yield high results with a relatively small amount of data and at a lower cost. The table below compares the characteristics of the two methods.
| Criteria | Fine-tuning | Embedding |
|---|---|---|
| Method of Knowledge Update | Adjust the entire model with a large amount of data | Retrieve and supplement necessary information from a pre-existing database |
| Performance Improvement | Performance increases if a large amount of similar data is available | Good performance if necessary information is well-organized |
| Required Resources | High-performance GPU, specialized knowledge, large dataset | Minimal dataset |
| Predictability of Behavior | Difficult to predict | Behavior is easier to predict because it’s explicit which data was used |
| Risk of Hallucination | High | Low |
| Cost | Relatively expensive ($0.008/1K words) | Relatively inexpensive ($0.06/1K words) |
- Hallucination
In the context of natural language processing, “hallucination” refers to the phenomenon where AI generates content that is either incorrect or irrelevant to the context. This is often the case with generative models and large-scale language models. This phenomenon can occur when the model generates “plausible” results based on the information from its training data.
Using GPT-4 for Summarizing Biobricks
GPT-4 is used to efficiently summarize data related to each Biobrick. The summarization was conducted using the following prompts. Also, a dataset of 550 parts from this year’s Asimov’s parts set was used.
Briefly summarise the information on the genetic part (biobrick).
What is this part used for? What function does it perform? What are the limitations and precautions for use?
Do not answer anything other than the question.
Show all in one paragraph.
Do not state which group made it or what year it was made, as it will be provided separately.
Data Details
Data for each Biobrick is managed in the following format:
- Registry Part ID: The ID registered in the registry.
- Short Name (unified): A unified short name.
- Registry Part: Detailed information about the part.
- Part Type (unified): The type of the part (terminator, cds, etc.).
- Registry Plasmid Backbone: The backbone of the plasmid.
Data Per Entry
Part ID BBa_J433041 Short Name L1 PGK-BFP positive control The BBa_J433041 part is a composite genetic element consisting of the Phosphoglycerate Kinase (PGK) promoter driving the expression of Blue Fluorescent Protein (BFP). Designed for mammalian systems, this part acts as a positive control to validate the PGK promoter’s ability to initiate gene expression. When functional, cells will display blue fluorescence, offering a straightforward method for both qualitative and quantitative evaluation of gene expression. Limitations include its mammalian-specific application and possible variations in fluorescence intensity depending on the cellular context. Precautions should be taken to ensure compatibility with the target mammalian system and to validate that fluorescence is an accurate representation of promoter activity. Part Type device Registry Plasmid Backbone BBa_J433100Asimov Asimov Mammalian Parts Collection(124 tokens= $0.00372)
In this manner, the system is designed in detail, and methods for future evaluations are also under consideration. Specifically, we are contemplating using various models to evaluate the quality of the answers provided.
4. Technology Stack Used and Reasons for Selection
Backend: Python 3
Python is widely used for various purposes including data science, web development, and automation. Specifically, it has a rich library for scientific computing and a large community, which is why it was chosen as the server-side language.
Database: Pickle
Pickle is Python’s standard serialization library that can save and restore the state of objects. In this project, Pickle was used to efficiently save and read data structures like vector indices.
Language Model: GPT-4
| Input | Output |
|---|---|
| $0.03 / 1K tokens | $0.06 / 1K tokens |
GPT-4 was selected because of its advanced natural language processing capabilities, which allow for high-quality dialogues and part recommendations. It is one of the latest models boasting high performance.
Embedding Model: Text-Embedding-Ada-002
| Usage |
|---|
| $0.0001 / 1K tokens |
This embedding model has the ability to convert natural language queries into vector form. It was chosen for this project due to its high performance and ease of code writing.
Frontend: JavaScript
JavaScript is one of the major programming languages used in web frontend development.
5. Recommendation Logic and Algorithm
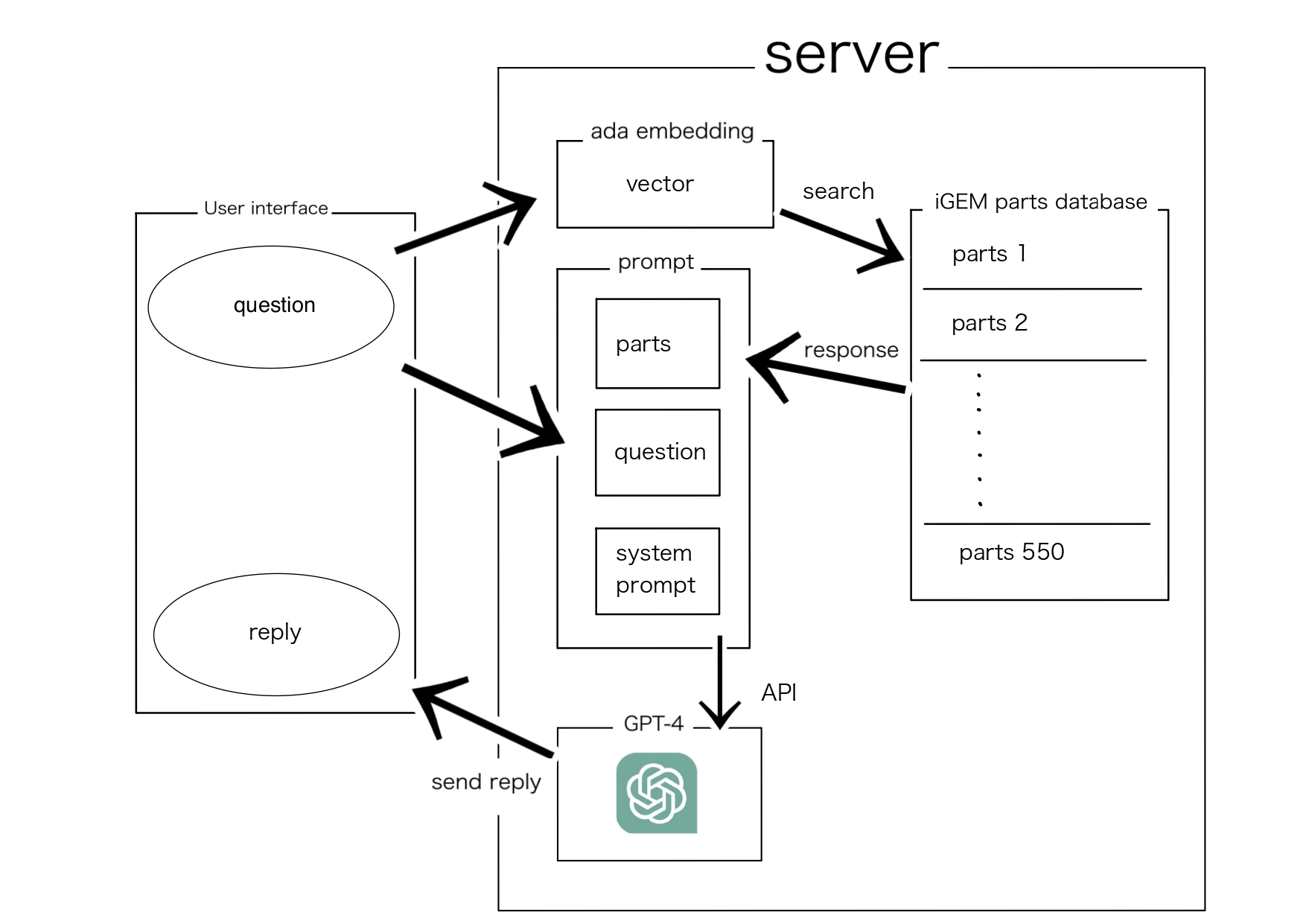
User Interface (UI):
Users access the web page and enter their questions or requests into a text box.
Frontend Processing:
Using JavaScript, the input from the user is captured and sent to the server.
API Endpoint Invocation:
The user’s questions or requests are converted into a vector (an array of numbers) by Text-Embedding-Ada-002. This is the query vector.
Database Query:
This query vector is compared with the vectors of each part in the database. Specifically, the degree of similarity is numerically evaluated using a calculation method called cosine similarity.
ChatGPT Processing Phase:
The part with the highest similarity and its information are sent to GPT-4. Based on this information, GPT-4 generates the most appropriate answer to the user’s question.
API Response:
The answer obtained from GPT-4 is sent to the frontend through the API.
Frontend Display:
The generated answer and the recommended part are displayed to the user.
Actual Response
- Example
command#1 I am working on combining multiple protein domains into a single polypeptide chain and am looking for genetic components to support this.
Selected Parts based on Similarity:
- Part ID BBa_J435223
- Part ID BBa_J435224
ANSWER: The genetic parts BBa_J435223 and BBa_J435224 could be useful for your project. These parts are designed for Yeast Two-Hybrid (Y2H) assays, which are used to discover protein-protein interactions.
BBa_J435223 encodes a fusion protein that combines the DNA binding domain of Gal4 with an activation domain (AD). This fusion protein can be used as the bait in a Y2H assay.
BBa_J435224 encodes the DNA-binding domain (BD) of the Gal4 protein and can be used as the prey fusion component in Y2H assays.
To combine multiple protein domains into a single polypeptide chain, you could design a construct where each protein domain is linked by a flexible linker sequence. This would allow each domain to fold and function independently. You could then use the Y2H assay to test whether the domains interact with each other in the context of the full-length polypeptide.
The success of this approach would depend on several factors, including the strength and specificity of the interactions between the protein domains, the expression levels of the fusion proteins, and the cellular environment.
command#2 I am searching for methods to efficiently integrate DNA into bacterial chromosomes. Specifically, I am considering chromosomal integration using one-step cloning techniques. I am looking for information on genetic parts or plasmids that support such techniques.
Selected Parts based on Similarity:
- Part ID BBa_J435115
- Part ID BBa_J433050
ANSWER: The genetic part BBa_J435115, also known as the pOSIP-CH (CamR, HK022) plasmid, is designed for one-step cloning and chromosomal integration of DNA. This plasmid was designed by Drew Endy and Keith Shearwin and facilitates the ease of integrating DNA into a host chromosome, which is a vital step in genetic engineering projects.
The pOSIP-CH plasmid contains the ccdB gene, and therefore, must be grown in a strain resistant to its product, like DB3.1. To repress the expression of the integrase, growth at 30°C is advised. However, the use of this plasmid requires the careful management of growth conditions and host strain selection due to the ccdB gene presence. It’s essential to follow the advised conditions to ensure successful DNA integration and to avoid potential toxic effects from ccdB expression.
Please note that the other given part, BBa_J433050, is a P1 process control plasmid under the control of the chicken elongation factor 1 alpha (chEF1a) promoter, part of the Asimov Mammalian Collection. This control plasmid is optimized for mammalian systems and may not be suitable for bacterial chromosomal integration.
command#2-2
What is the DB3.1?
ANSWER:
DB3.1 is a strain of Escherichia coli that is commonly used in molecular biology, particularly in the cloning of DNA. It is a chemically competent cell line, meaning it can easily take up foreign DNA, such as plasmids. This makes it useful for genetic engineering and biotechnology applications.
command#3 What are Biobricks for knocking down specific genes using the CRISPR/Cas9 system?
ANSWER:
The part BBa_J428062 is a variant of hSpCas9, a DNA-targeting endonuclease used in CRISPR systems. This part is used for a variety of genome editing applications including gene knockout, gene insertion, and gene regulation. Users interested in DNA editing should consider this part and may need to validate its functionality for their specific needs. This part is compatible with standard assembly methods and is part of the CRISPR-Cas Collection in the iGEM Registry.
However, it’s important to note that to knock down specific genes using the CRISPR/Cas9 system, you would also need guide RNAs (gRNAs) that are designed to target the specific genes of interest. These gRNAs are not included in the BBa_J428062 part and would need to be designed and synthesized separately.
command#4 What Biobricks are suitable for highly efficient gene expression in Escherichia coli?
ANSWER:
The given parts include BBa_J23110, a constitutive promoter known for medium-level strength in gene expression, and BBa_J435302, a ribozyme designed to reduce DNA context effects on gene expression. Both parts have been engineered for use in Escherichia coli.
BBa_J23110 has been extensively used and characterized, with successful cloning into E. coli and non-model organisms like agrobacterium and rhizobium. It has been reported to drive gene expression effectively, with fluorescence levels nearly fifteen times above the baseline in E. coli. However, it’s worth noting that the expression level can vary depending on the gene and E. coli strain used. Also, stronger Anderson Series promoters, including J23110, have been found to impose a metabolic burden on E. coli, affecting growth rates.
BBa_J435302 aims to make expression levels more predictable by minimizing the influence of surrounding DNA sequences. It is flanked by 5’ and 3’ fusion sites to facilitate easier cloning and integration into expression vectors.
command#5 What is the best way to inactivate pathogens caused by bacteria?
ANSWER: The best way to inactivate pathogens caused by bacteria would typically involve a combination of strategies, including the use of antibiotics, disinfectants, and potentially bacteriophages. However, based on the given parts, we can propose a more specific strategy.
The part BBa_J435096 encodes for beta-lactamase, an enzyme that cleaves a wide range of beta-lactam compounds, typically antibiotics. This enzyme could potentially be used to inactivate beta-lactam antibiotics produced by certain bacteria, thereby reducing their ability to kill other bacteria and allowing for a more diverse bacterial population that could outcompete the pathogens.
Additionally, the parts BBa_J435054 and BBa_J435044, which encode for Endonuclease VIII and IV from E. coli respectively, could potentially be used to damage the DNA of the pathogens. These enzymes are involved in DNA repair processes, but if overexpressed, they could potentially cause excessive DNA damage and lead to cell death.
However, it’s important to note that these strategies would require careful control and regulation to ensure that they are specifically targeting the pathogenic bacteria and not causing harm to beneficial bacteria or the host organism. Furthermore, these strategies would likely need to be combined with other methods, such as the use of antibiotics or other antimicrobial agents, to effectively inactivate the pathogens.
command#6 I want to create bacteria that produce insulin with high efficiency. Which parts do you recommend?
ANSWER: For creating bacteria that produce insulin with high efficiency, you may consider the following parts:
BBa_J435217 (Cre): This part can be used for facilitating Cre-LoxP recombination in your bacterial system. This can be useful for manipulating gene arrangements within the genome, such as gene insertion of the insulin gene.
BBa_J428041 (pL_lac0_1): This is a strong promoter that is repressible by both LacI and Lambda-CI proteins. It can be used to control the expression of the insulin gene. Given its strong activity, it would be ideal for applications where high expression levels are needed.
BBa_J435330 (AF_high copy (pUC) ori/KanR): This part contains a high-copy-number origin of replication along with a kanamycin resistance gene. This allows for high-level amplification of the plasmid and provides resistance to kanamycin for selection purposes. The high-copy-number feature facilitates higher yields of plasmid DNA, which can be beneficial for high-level insulin production.
BBa_J23110: This is a constitutive promoter known for medium-level strength in gene expression. It can be used as an alternative to the pL_lac0_1 promoter for driving the expression of the insulin gene, depending on the specific needs and conditions of your project.
Please note that the effectiveness and efficiency of these parts can be influenced by various factors, such as the specific strain of bacteria used, the growth conditions, and the overall design of your genetic circuit. Always consider these factors when selecting and using these or any other genetic parts
command#7 Are there Biobricks that control communication between bacteria?
ANSWER: Yes, there is a Biobrick that controls communication between bacteria. The BBa_C0070 part encodes for the RhlI protein, an autoinducer synthetase that produces N-butyryl-HSL (BHL). This molecule is a signaling compound that specifically binds to the RhlR protein, initiating quorum-sensing mechanisms. Sourced from Pseudomonas aeruginosa, this part is primarily used to generate BHL for quorum sensing studies or synthetic biology applications requiring controlled gene expression.
command#8 Is there a way to store data using bacterial DNA?
ANSWER: Yes, it is theoretically possible to store data using bacterial DNA. The process would involve encoding the data into a sequence of DNA bases (A, T, C, G), synthesizing this DNA sequence, and then integrating it into the bacterial genome or a plasmid. The data could then be retrieved by sequencing the DNA and decoding the sequence back into its original format.
In the context of the given parts, the BBa_J435053 (T4 g32) could potentially be used to stabilize the synthesized DNA during the encoding and decoding processes. The BBa_J435251 (Pichia PARS) could serve as a plasmid for storing the data, as it is designed to function as an episome within the cell, providing a separate, self-replicating genetic element.
The BBa_J433047 (L2 2xTUs - lacZ) could be used as a marker for selection, allowing for the identification of successfully transformed cells that have incorporated the data-storing DNA.
However, it’s important to note that this is a highly simplified explanation and the actual process would be much more complex and would require a detailed understanding of genetic engineering and bioinformatics. Additionally, while DNA data storage is a promising area of research, it is still in its early stages and there are many technical challenges that need to be overcome.
command#9 Is there a way to make environmentally friendly dyes using bacteria?
ANSWER: Yes, it is possible to make environmentally friendly dyes using bacteria. The given parts provide a variety of chromoproteins that can be used as colorimetric reporters, producing colors visible to the naked eye. These proteins can be expressed in bacteria such as E. coli to produce color, essentially acting as biological dyes.
For example, the part BBa_K592011, known as cjBlue, produces a dark green color when expressed in E. coli. Similarly, BBa_J435081, also known as gfasPurple/gfasCP, produces a color visible to the naked eye. BBa_K1033919, known as gfasPurple, produces a visibly purple color. BBa_J435104 is another variant of cjBlue that can be used as a colorimetric reporter. BBa_K864401, known as aeBlue, produces a deep blue color. Lastly, BBa_K592100 encodes the Blue Fluorescent Protein (mTagBFP), which fluoresces blue under specific conditions.
These parts can be introduced into bacteria using standard genetic engineering techniques. Once inside the bacteria, they can be expressed to produce the corresponding proteins, which in turn produce the desired color. The bacteria can then be grown in large quantities to produce the dye. This approach is environmentally friendly as it does not require the use of harmful chemicals typically used in dye production. Furthermore, the color produced by these proteins is stable and does not fade easily, making them suitable for use as dyes.
command#10 I want to discover bacteria adapted to a particular environment. What can I use?
ANSWER: The given parts do not directly provide a tool for discovering bacteria adapted to a particular environment. However, they can be used in a roundabout way to achieve this.
For instance, you could use the antibiotic resistance markers (BBa_J435277, BBa_J435278, BBa_J435257, BBa_J435280) to create a selection pressure in a culture of bacteria from the environment of interest. By introducing these markers into the bacteria and then applying the corresponding antibiotic, you could select for bacteria that have successfully incorporated the marker, which might indicate a higher level of adaptability or competence.
The part BBa_J435006, which encodes the EcoRI restriction enzyme, could be used to cut DNA at specific sites, facilitating the insertion of the antibiotic resistance markers into the bacterial DNA.
However, this approach would only select for bacteria that are competent in taking up and incorporating foreign DNA, and not necessarily bacteria that are best adapted to the environment. Additionally, this approach requires a priori knowledge of the bacteria’s genetic makeup and the ability to genetically manipulate them, which may not be feasible for all environmental bacteria.
In conclusion, while the given parts can be used in creative ways to study bacterial adaptability, they are not directly suited for discovering bacteria adapted to a particular environment.
Evaluating the dataset and assessing the quality of the answers are still challenges. We are considering using models to evaluate the quality of the responses. For reference, see LLM-Eval.
6. Discussion
Challenges Encountered and Solutions
There were several difficulties in the early stages of the project. The most notable was that users experienced long wait times. As a solution to this problem, we introduced streaming mode. This allowed text to be displayed to the user almost instantly, character by character.
Next, we encountered challenges when vectorizing genetic parts based on queries in natural language. The simple combination of vectorization and cosine distance was not sufficient to identify the most appropriate parts. To solve this issue, we considered the introduction of a hybrid approach that combines vector search technology with keyword search.
Future Possibilities
To improve the quality of part recommendations, having a database with information on the background and actual use-cases for each part could enable more detailed part recommendations.
In the future, we are considering introducing locally-operating LLMs (e.g., Xwin-LM) to improve the system’s cost-efficiency and reproducibility. Overall, the system is still under development, and all its potentialities are unexplored. We plan to continually improve and expand the system while utilizing feedback from the user community. We aim to deepen the collaboration between future iGEM and LLM technologies.
Ultimately, we aim for Parts-LLM + model support LLM to automatically select the target parts, set up the model, store the experimental notes, and output them to a wiki using a multi-modal LLM.
Additionally, this information is as of October 9, 2023, and details such as the pricing of GPT APIs may differ from the current situation.